Jupyter notebook is one of the most favorite tools of data science professionals; however, developing everything in a Jupyter notebook can be error prone. In addition, even though there are workarounds, git itself does not track Jupyter notebooks. That being said, a Jupyter notebook can certainly be helpful to specific parts of a data science project, including exploratory data analysis, which would require a lot of experimentation and visualization. Today, we want to explore an option offered by VSCode that combines both the advantage of using a Jupyter kernel and using a .py file.
How to Use It:
First, install the ipykernel library within a virtual environment.
(venv) pip install ipykernel
Then type in # %% in a .py file within your VSCode is all you need to activate the Jupyter kernel within the .py file, which will create a code block just like a Jupyter Notebook.
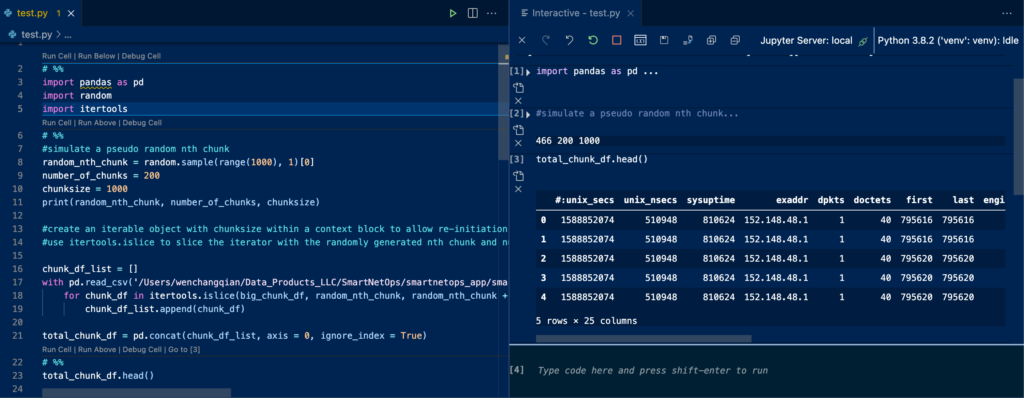
Then press shift+enter to execute the code block to see the code output from the right side. To make sure you are using the right kernel, you can type the following to see which python you are using.
! which python
Separating the Testing Code and Development Code:
Even though this is not a total separation of kernel environment, it’s still a better practice to separate your testing code and development code. It’s very easy to rename the same variables or delete a block of code which contains a variable name that you ended up using later within a Jupyter notebook; those all can potentially create bugs in your notebook. You can still explore the data and develop visualization on the right side of the IDE, however obviously you don’t have the capability of saving the statistical/visualization output with the introduced method, which a Jupyter notebook would give you.
Faster Development Speed and Better Coding Habit:
A Jupyter notebook allows you to see results for each coding block, which may sometimes allow new developers to form habits of writing non-pythonic code or one-liners to just get the results. However, enforcing yourself to compile large blocks of code, i.e. a class like the following, in your head when you write it may help you to design better classes and develop faster. The longer you do this, the more capable you might be to develop longer low-bug code without spending much time on testing along the way. This also reduces the time to extract the code or refactor the code into functions/classes from your Jupyter notebooks, if you didn’t do that within a notebook.
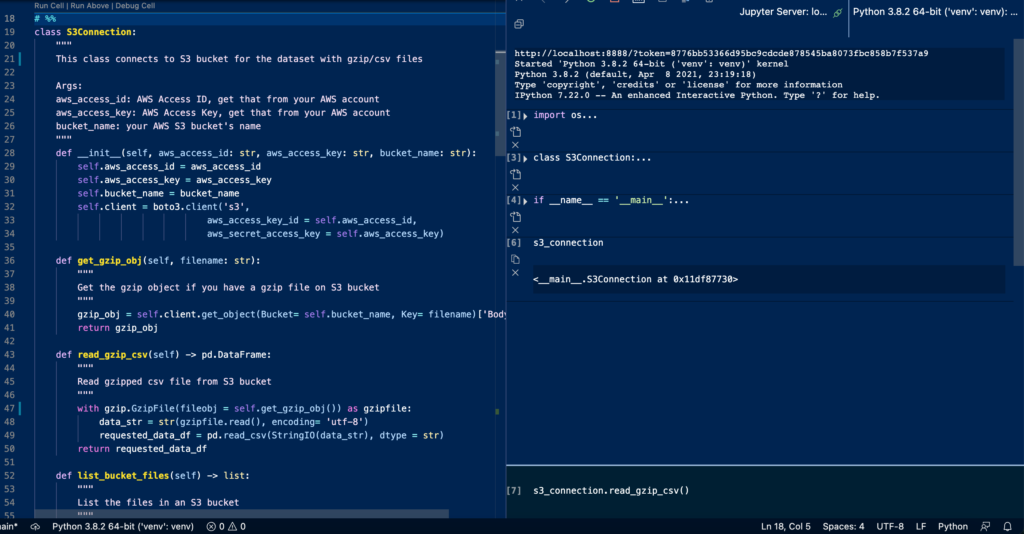
More Distributable and Better Versioning
A lot of times, data science professionals don’t just work by themselves as they would have to collaborate with other engineering professionals. Being able to keep all the code or most of the code base in .py files will make a data science professional’s work more distributable and easier to engineer. On the other hand, git has trouble versioning and tracking .ipynb files. Although there are different ways to work around it, they do add a little bit more complexity to the project and some of them are paid options with free tiers (public repos only).
That’s it for today’s data hack. Hope you like it!